Biophysics of protein folding
During the protein folding process, the unfolded protein chain converts into the native, biologically active conformation. In this context, we investigate the unfolded state and folding intermediates in equilibrium with the native state as well as protein folding kinetics by NMR, circular dichroism, and fluorescence spectroscopy.
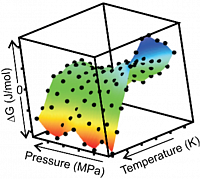
Stability profile of protein Kti11 depending on temperature and pressure determined by NMR.
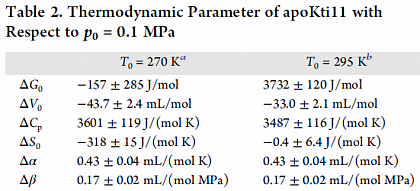
Thermodynamic parameters derived from the stability profile.
We put special emphasis on the characterization of transient folding intermediates. For the human CDK inhibitor p19INK4d and the gene 3 protein from fd phage we could show that transient states are directly related to function. E.g. the regulation of the CDK 4/6 inhibition during the human cell cycle by p19INK4d occurs via its five ankyrin repeats of different stability revealed from structural studies of folding intermediates. Another example is the infectious state of the gen 3 protein, which is not most the stable one but a folding intermediate.
The structural biology of protein reactions can be investigated by time-resolved NMR, because we are working in solution.
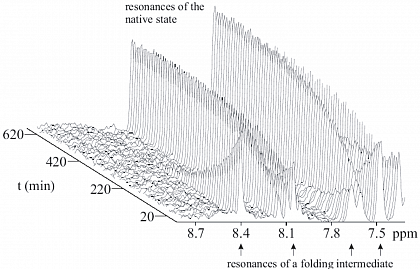
Refolding kinetics of an folding intermediate and the native state of the protein ribonuclease T1.

