Research Ulrich Weininger
My research interests are structures and dynamics of proteins in general, and the dynamic of amino acid side chains in particular, with a focus on aromatic side chains.
NMR structures of proteins
The following protein structures have been solved by me or with my help using NMR spectroscopy:
methods for the study of side chain dynamics
Following methods have been developed for the study of side chain dynamics:
|
|
isotope labeling |
site selective 13C labeling of proteins using erythrose
site selective 13C labeling of proteins using ribose
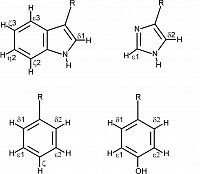
isotope labeling of aromatic side chains
|
|
NMR dynamics of methyl groups |
1H and 13C CPMG relaxation dispersions of methionine
1H R1rho relaxation dispersions of methyl groups

|
|
NMR dynamics in aromatic side chains |
13C relaxation experiments in aromatic side chains
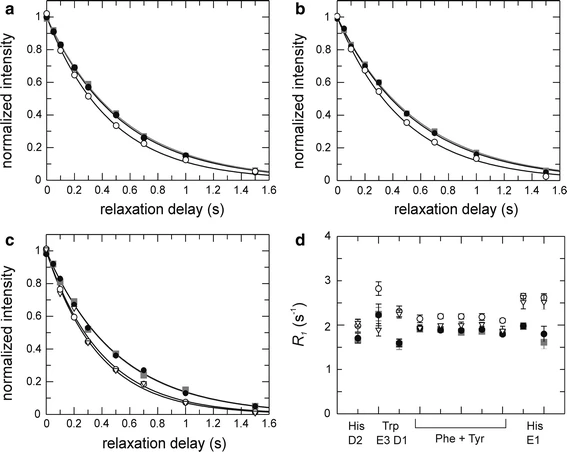
13C CPMG relaxation dispersions in aromatic side chains
1H CPMG relaxation dispersions in aromatic side chains
1H CPMG relaxation dispersions in aromatic side chains 2

13C R1rho relaxation dispersions in aromatic side chains
1H R1rho relaxation dispersions in aromatic side chains
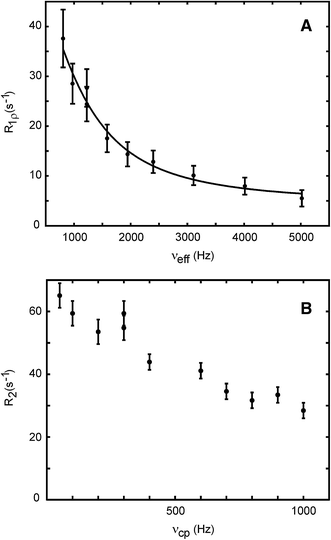
RDC mediated relaxation dispersions in aromatic side chains

side chain dynamics in proteins
research of general function of proteins based on side chain dynamics
dynamics of aromatic side chains in the active site of FKBP12

dynamic allosteric communication in the glucocorticoid receptor

energetics and dynamics of the proton shuttle of CA II
ring flips
experiments on aromatic side chain dynamics enable the identification and quantification of ring flips, which report on transient rearrangements in proteins
NMR studies of ring flips

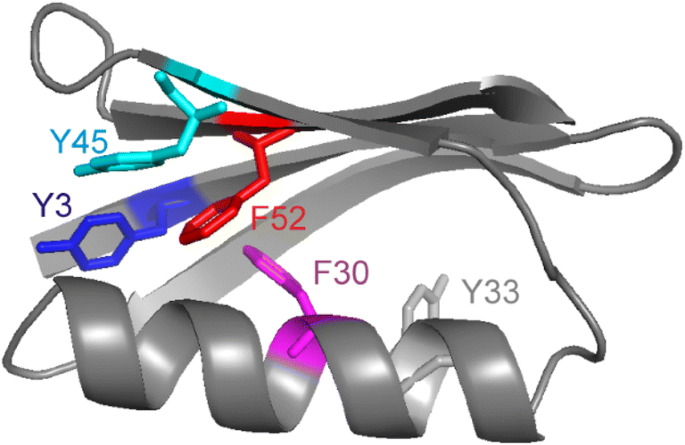
ring flips in BPTI revisited

ring flips in the aromatic cluster of GB1
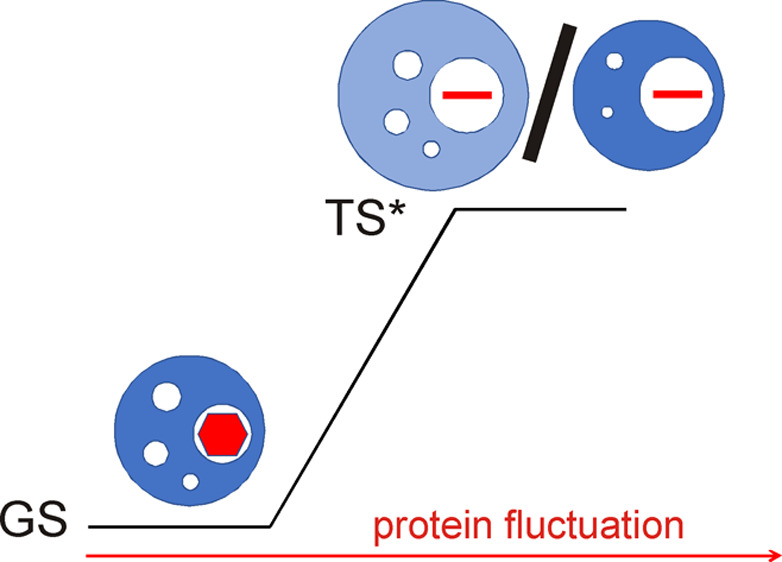
ring flips despite no chemical shift difference

transition state compressibility of ring flips
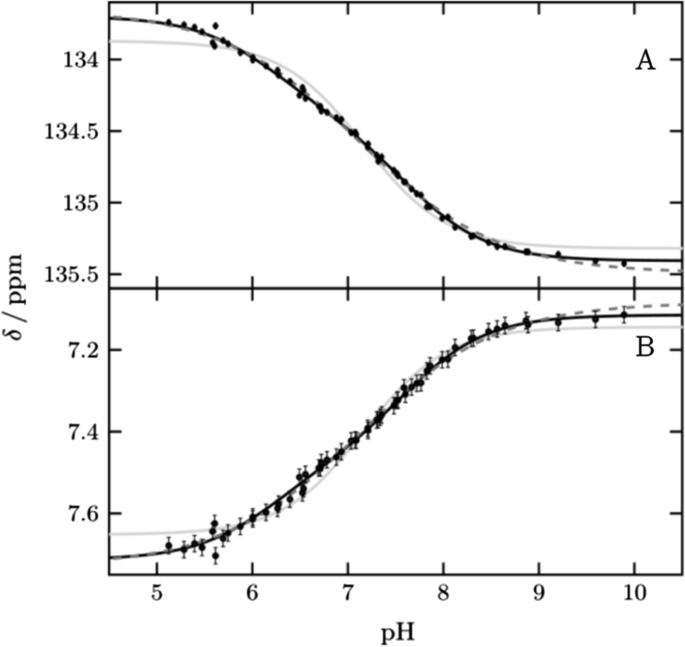
protonation kinetics of ionizable side chains
NMR relaxation enables to meassure the protonation kinetics (on & off) in ionizable side chains of proteins in a pH and pKa dependent manner
site-specific protonation kinetics of acidic side chains

proton transfer kinetics in histidine side chains

protonation states of histidine
histidine can exist in a combination of 3 states, which can be investigated by NMR
the carbohydrate-binding site in Gal3 is preorganized
proton occupancies in histidine side chains

pKa value determination
experimental pKa value determination of a hyperstable protein
energetics and dynamics of the proton shuttle of CA II
proton occupancies in histidine side chains

aggregation
coaggregation of Ab40 and Ab42
DNAJB6 inhibits the aggregation of Ab42
alpha synuclein aggregation
sekundary nucleation

protein folding by NMR
stabilization of the native state of by pressure
protein folding by NMR
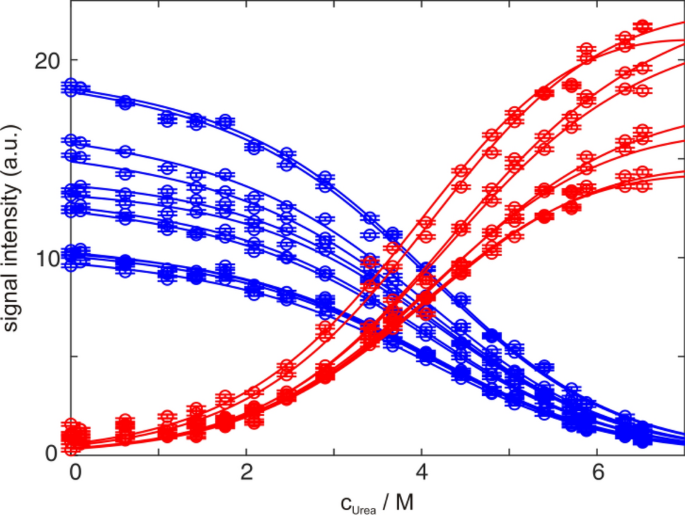
PPIases
substrate recognition and catalytic mechanism of SlyD
impact of distant peptide substrate residues on enzymatic activity of SlyD