Structure determination of proteins
NMR spectroscopy allows the determination of the three dimensional structures of biomacromolecules at high resolution. Thousands of distances between 0.2 - 0.5 nm, many bond angles and relative bond orientations are experimental constraints for molecular dynamics calculations of the protein structure.
For the following proteins, we could determine the NMR structure: cold shock protein CspB from Bacillus subtilis (2F52.pdb), parvulin domain PpiD from Eschericia coli (2KGJ.pdb), plasmid copy control protein ORF56 from Sulfolobus islandicus (2K9I.pdb), prolyl isomerase and molecular chaperons SlyD (2K8I.pdb) and SlpA (2M2A.pdb), helix-0 of human N-BAR in SDS (2RMY.pdb) and DPC micelles (2RND.pdb), auxin transcription faktor IAA4 from Pisum Sativum (2M1M.pdb).
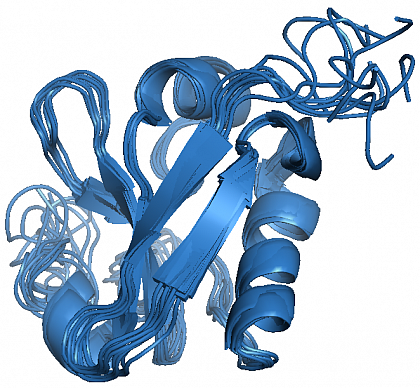
NMR structure of PsIAA4 (ensemble of the 9 lowest energy structures).
Together with colleagues the following protein structures could be determined by X-ray crystallography: cold shock protein and complexes with nucleic acids (2ES2.pdb, 2HAX.pdb, 3PF4.pdb, 3PF5.pdb), ankyrin repeat protein tANK from Thermoplasma volcanium (2RFM.pdb), prolyl isomerase and molecular chaperone SlyD from Thermus thermophilus (3CGM.pdb, 3CGN.pdb, 3LUO.pdb), plasmid copy control protein ORF56 from Sulfolobus islandicus (3FT7.pdb), zinc-finger containing Kti11 From Saccharomcyes cerevisiae (5AX2.pdb), onconase C87A/C104A From Rana pipiens (3HG6.pdb), pilus and TolA binding domain of IF1 und IKe phage (4EO0.pdb, 4EO1.pdb, 2X9A.pdb, 2X9B.pdb, 3KNQ.pdb).
Structural investigations of the human Bin1 N-BAR protein, which is involved in membrane remodeling, could be achieved in collaborations by cryo-electron microscopy.

